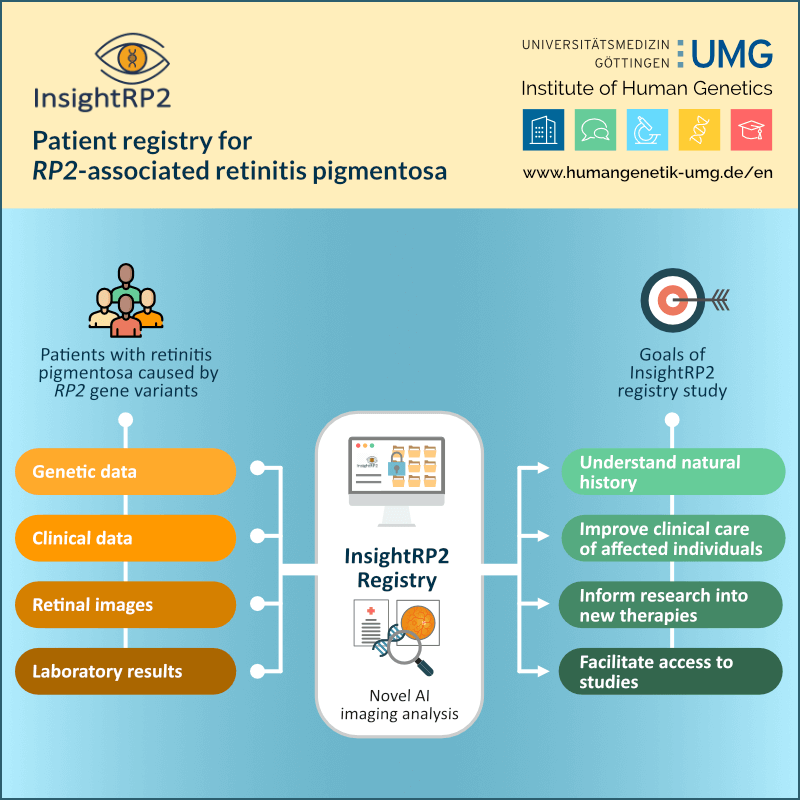
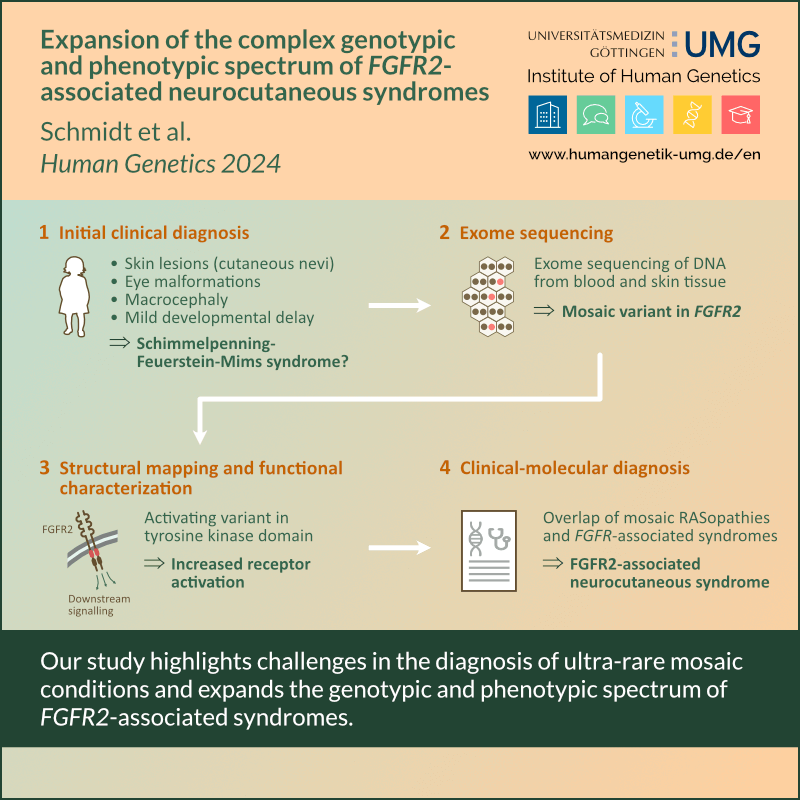
Identified heterozygous truncating SEC24C variants affect protein transport and glycosylation and cause previously undescribed syndrome with epilepsy, cataracts and anemia
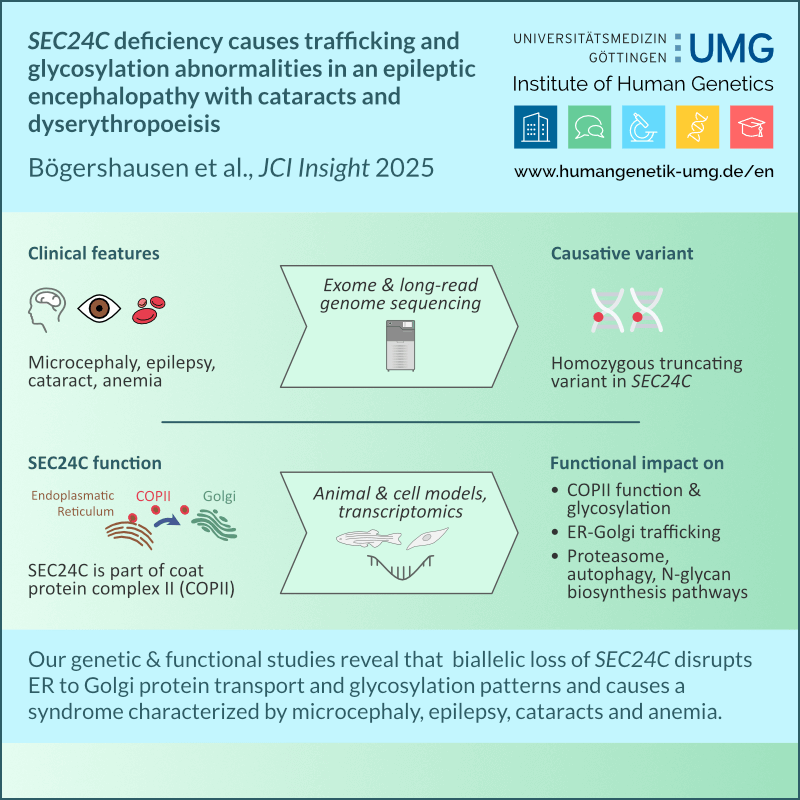
SEC24C deficiency causes trafficking and glycosylation abnormalities in an epileptic encephalopathy with cataracts and dyserythropoeisis
Bögershausen N, Cavdarli B, Nagai T, Milev MP, Wolff A, Mehranfar M, Schmidt J, Choudhary D, Gutiérrez-Gutiérrez Ó, Cyganek L, Saint-Dic D, Zibat A, Köhrer K, Wollenweber TE, Wieczorek D, Altmüller J, Borodina T, Kaçar D, Haliloğlu G, Li Y, Thiel C, Sacher M, Knapik EW, Yigit G, Wollnik B.
JCI Insight. 2025 Mar 25:e173484. doi: 10.1172/jci.insight.173484. Epub ahead of print.